Data Querying + Gen3 SDK¶
Overview ⚙️¶
Gen3 supports API access to Files and Metadata, allowing users to download and query their data via the Gen3 SDK and GraphQL queries.
Quick Start ⚡️¶
Adapted from the Gen3 SDK Quick Start page
1. Dependency and Credentials¶
Prior to installing, check a profile credentials. Test:
- will return a list of projects that a profile has access to.-
For new setup or renew of gen3 credentials - Follow steps to configure/re-configure a profile with credentials:
- Download an API Key from the Profile page and save it to
~/.gen3/credentials.json
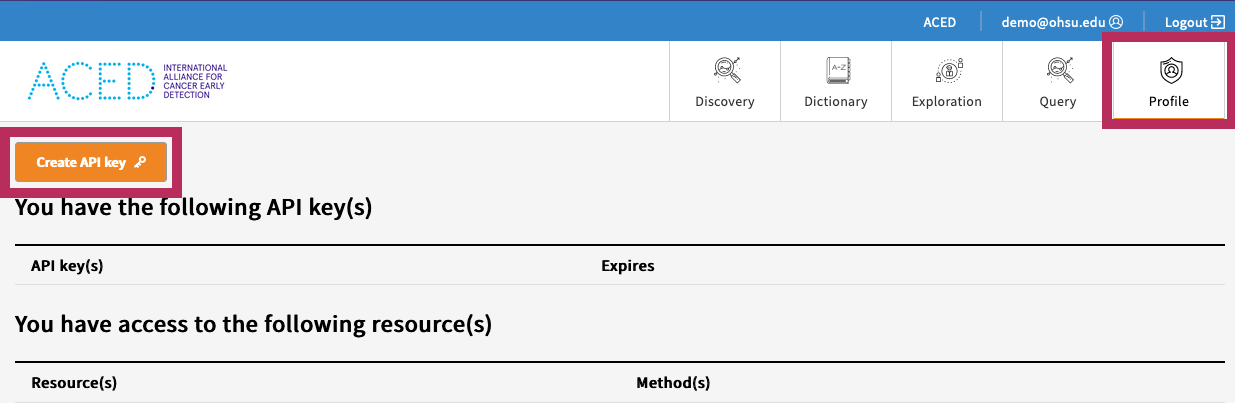
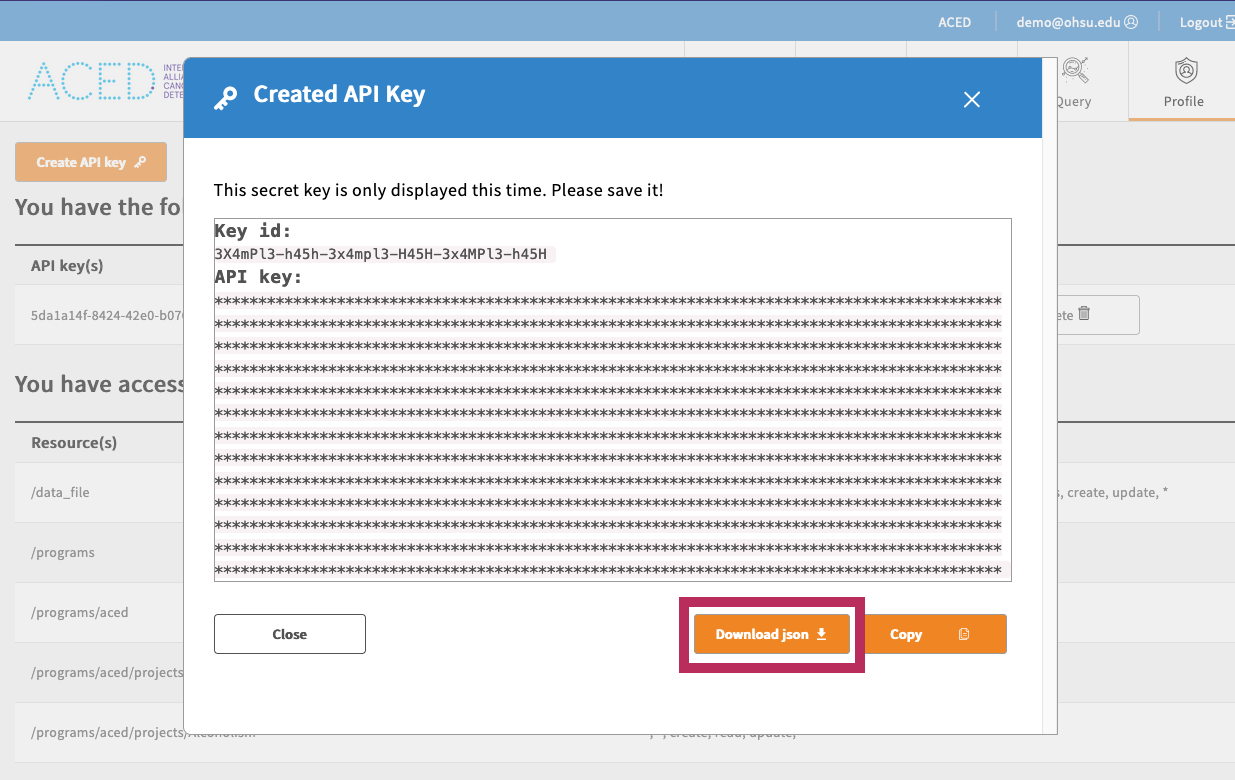
- Download an API Key from the Profile page and save it to
2. Install¶
The Gen3 SDK is available for installation via PyPi:
3. Query¶
The following query examples provide a high-level overview on how to use Gen3 python SDK to authenticate, use the authentication to fetch column/metadata names, retrieve entity ids, and then use the ids to make nested queries. Each project would have a graph schema or definition with relations associated that are defined during data transformation or harmonization. The queries would depend on each project's graph definition traversals.
3.0 Authenticate¶
3.1 List available fields on an entity to query on¶
def get_entity_fields(entity_name, auth):
"""Retrieve all field names for a given entity from the Gen3 GraphQL schema."""
query_template = f"""
{{
__type(name: "{entity_name}") {{
fields {{
name
}}
}}
}}
"""
response = Gen3Query.graphql_query(
Gen3Query(auth),
query_string=query_template
)
if response and response.get("data", {}).get("__type", {}):
fields = response["data"]["__type"]["fields"]
field_names = [field["name"] for field in fields]
return field_names
else:
print(f"Failed to retrieve fields for {entity_name}:", response)
return []
entity_name = "Specimen"
field_names = get_entity_fields(entity_name, auth)
print(f"Available fields for {entity_name}:", field_names)
3.2 Filter Patients of Interest (ResearchSubject Participants)¶
researchsubject_query = """
query ($filter: JSON) {
researchsubject(filter: $filter, first: 10000) {
id
patient_id
condition_diagnosis
}
}
"""
researchsubject_variables = {
"filter": {
"IN": {
"condition_diagnosis": ["Infiltrating duct carcinoma, NOS"]
}
}
}
researchsubject_response = Gen3Query.graphql_query(
Gen3Query(auth),
query_string=researchsubject_query,
variables=researchsubject_variables
)
patient_ids = [p["patient_id"] for p in researchsubject_response["data"]["researchsubject"]]
3.3 Filter Specimens of Interest from Patient Ids¶
def execute_query(auth, query_string, variables=None):
"""perform guppy query"""
return Gen3Query.graphql_query(Gen3Query(auth), query_string=query_string, variables=variables)
def get_specimens(auth, patient_ids):
"""Fetch specimens associated with patient ids"""
query = """
query ($filter: JSON) {
specimen (filter: $filter, first: 10000) {
id
patient_id
specimen_id
<other_sample_metadata>
}
}
"""
variables = {"filter": {"IN": {"patient_id": patient_ids}}}
response = execute_query(auth, query, variables)
specimens = defaultdict(list)
for specimen in response["data"]["specimen"]:
if specimen["gene"]:
specimens[specimen["patient_id"]].append(specimen)
return specimens
dat = get_specimens(auth, patient_ids)
specimen_ids = [d[1][0]['id'] for d in dat.items()]
3.4 Filter Files Associated with Specimens of Interest¶
def get_files(auth, specimen_ids):
"""Fetch files associated with specimens."""
query = """
query ($filter: JSON) {
file (filter: $filter, first: 10000) {
id
specimen_id
sample_type
}
}
"""
"""
variables = {
"filter": {
"AND": [
{
"IN": {
"specimen_id": specimen_ids
}
},
{
"LIKE": {
"sample_type": ["%Primary Tumor%"]
}
}
]
}}
"""
variables = {"filter": {"IN": {"specimen_id": specimen_ids}}}
response = execute_query(auth, query, variables)
return response["data"]["file"]
file_data = get_files(auth, specimen_ids)
Simple end to end workflow:¶
Query (example.graphql)¶
query ExampleQuery {
files: file(first: 1000) {
file_name
project_id
id
}
patients: patient(first: 1000) {
name
project_id
id
}
observations: observation(first: 1000) {
code
project_id
id
}
}
Script (example.py)¶
from gen3.auth import Gen3Auth
from gen3.query import Gen3Query
import json
auth = Gen3Auth()
query = ''
# Read in Example Query
with open('example.graphql') as f:
query = f.read()
response = Gen3Query.graphql_query(Gen3Query(auth), query_string=query)
formatted = json.dumps(response, indent=2)
print(formatted)
# >>> Example Output
Output¶
$ python example.py
{
"data": {
"files": [
{
"file_name": "example.bam",
"project_id": "cbds-example",
},...
],
"patients": [
{
"name": "Example Name",
"project_id": "cbds-example",
},...
],
"observations": [
{
"code": "Example Code",
"project_id": "cbds-example",
},...
]
}
}